Find Links Between Drugs, Genes, Diseases, and Biological Pathways
The Dataiku Solution for Drug Repurposing Knowledge Graph provides fully clean and blended datasets in the form of nodes and relationships leveraging numerous public sources of data. Embrace the potential of graph analytics for drug repurposing.
Ease Ingesting and Blending Data
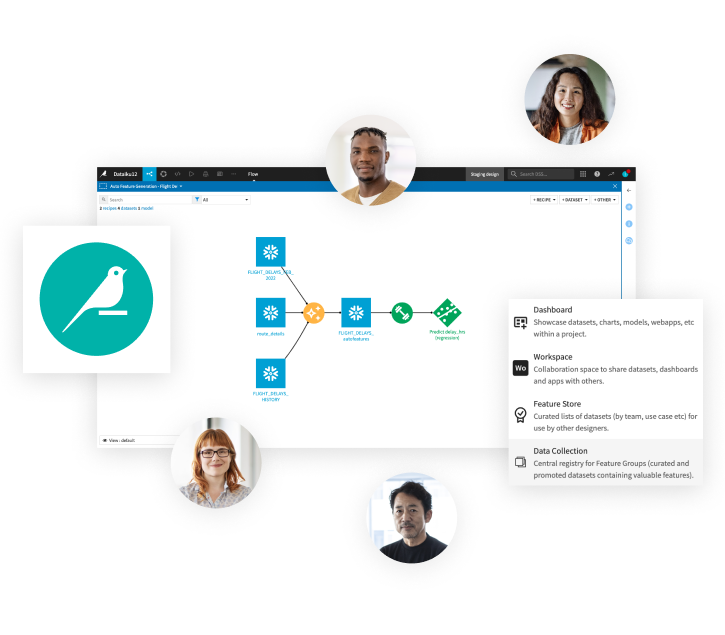
Make ingesting, cleaning, and blending of all relevant data easier with the solution’s clear flow zones and full wiki. Transform external data into a network graph ready to export to a graph database or leverage to enrich your biological research and development analysis projects.
Keep Workflows
Up to Date
Automate ingestion of key public data sources on drugs, diseases, and genes through FTP and Postgres from NCBI, DrugCentral, and related gene ontologies and pathway databases. A variety of FTP connectors allow for single-click refresh and rebuild. Automate these downloads to keep your knowledge graph current to new discoveries and biological findings.
Identify Novel Patterns and New Indications Easily
Create relationships between nodes in your data leveraging Dataiku’s Visual Graph Editor plugin. Network graphs can be created in an entirely visual manner to test the strength of node/edge relationships with no need for coding.
Dive Into Complex Relationships With a Biomedical Knowledge Graph
Push final graphs in neo4J for further investigation and use in various day-to-day activities.
Go Further With
Graph Analytics
Create scenarios for new data ingestion and run queries to improve biological relationship understanding. Start with open data and deepen your nodes with more data sources. Embed graph analytics into your data-driven strategies to maximize complex relationship identification. Share biological and drug chemistry relationship data sets with other analytics and machine learning projects such as molecular structure property prediction and drug safety outcomes research.
Answer Key Questions on Drugs Development With Graph Analytics
The Dataiku Solution for Drug Repurposing Knowledge Graph helps answer a broad range of questions like:
- How can I more efficiently find links between drugs and potential new uses?
- How can I work with various sources of data like MeSH, Gene Ontology, Uberon, Pathway Commons, and DrugCentral?
- How can I keep my datasets current with new information and developments from the scientific community?
- How can I convert structured datasets into network graphs without knowing complex coding?
Knowledge Graphs Are Just the Start
Tap into the full breadth of potential offered by Dataiku to enrich your day to day R&D activities with accelerated usage of data. From simplified data access and analysis down to complex ML-assisted modeling, embrace the potential of data with a fully governed approach.
The Total Economic Impact™️
Of Dataiku
A composite organization in the commissioned study conducted by Forrester Consulting on behalf of Dataiku saw the following benefits:
reduction in time spent on data analysis, extraction, and preparation.
reduction in time spent on model lifecycle activities (training, deployment, and monitoring).
return on investment
net present value over three years.